2. Chemical biology
In our synthetic chemistry research, we typically utilize purified simple substrates to achieve high yield and selectivity by optimizing reaction conditions through the careful adjustment of catalysts, reagents, solvents, and reaction temperatures. However, mixtures derived from living cells are far from these ideal, purified conditions, so artificially realizing site-selective modification under physiological conditions presents many new challenges. In nature, enzymes provide a solution. Enzymes facilitate various bond-forming and bond-cleaving reactions with remarkable selectivity, even amidst numerous biomolecules that contain inherently reactive functional groups. Our research aims to bridge the gap between chemistry and biology by developing new synthetic strategies, with a focus on protein methylation as a model study.
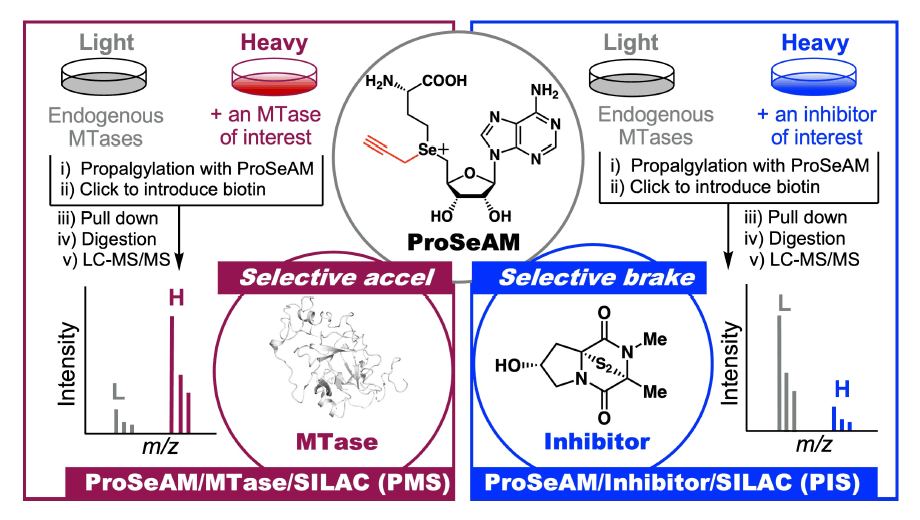
2-1) Development of probes: Visualize (PMS: ProSeAM/MTase/SILAC)
Protein methylation reactions are crucial post-translational modifications that regulate various biological functions. Despite their importance, only a small fraction of these methylation events have been identified in living cells. To further map methylation in living cells, we are integrating chemical and biological technologies. Our strategy involves introducing an artificial tag into native protein substrates using a designed derivative of the natural methyl donor S-adenosylmethionine (SAM). Notably, we discovered that ProSeAM (propargylic Se-adenosyl-L-selenomethionine) is effective for tagging native protein substrates with a propargyl functional group. Moreover, we found that adding exogenous recombinant methyltransferase (MTase), a natural catalyst, enhances the production of the corresponding propargylated product even in cell lysates. By combining this approach with the SILAC (Stable Isotope Labeling by Amino acids in Cell culture) comparative quantitative method, we have developed a technique that allows for highly accurate correlation of enzyme and substrate within the same chromatogram. This versatile method has already successfully identified several poorly characterized non-histone substrates and their modification sites.
2-2) Development of inhibitors: Control (PIS: ProSeAM/Inhibitor/SILAC)
In traditional research on the development of protein methylation inhibitors, the optimization of inhibitor structures is guided by the methylation reaction of a "one enzyme–one substrate–one site" model. In contrast, we have developed the ProSeAM/Inhibitor/SILAC (PIS) method, which utilizes the ProSeAM detection system to evaluate the inhibitory activity of small molecules of interest. By using the natural product chaetocin as a seed compound, we have successfully created novel arginine methylation inhibitors with reduced toxicity.
Reviews: Acc. Chem. Res. 2021, 54, 3818–3827; Chem. Rec. 2018, 18, 1660–1671.
Representative papers: PLOS ONE, 2014, 9, e105394; Chem. Commun. 2018, 54, 9202–9205; Nat. Commun. 2021, 12, 891; Genes. Dev. 2023, 37, 724–742.